5 + 5[1] 10Week 11 – lecture A
November 1, 2025
This is the first of four-weeks series that will focus on the R language. Besides being a great all-round tool for data analysis and visualization, the R language is probably the best environment for the “downstream” analysis of omics data, with an excellent array of packages for e.g.:
Specifically, you will learn:
In this lecture, you will learn:
You can use R with a “console” that can simply be run inside the terminal (at OSC, you can do so after loading the R module). However, we’ll use R with a fancy editor (or “IDE”, Integrated Development Environment) called RStudio, which makes working with it much more pleasant!
Similar to VS Code, we can run a version of RStudio (“RStudio Server”) within our browsers by starting an Interactive App through OSC OnDemand.
Interactive Apps at the bottom, click RStudio Serverpitzer4.4.0PAS28802any1LaunchConnect to RStudio Server button once it appearsNote that unlike with Code Server, we can choose how many cores we want when starting an RStudio server job. Changing the number of cores is also the way to go if you need more than the default 4 GB of memory (RAM): recall that each core is associated with 4 GB of memory, so asking for 4 cores will get you 16 GB RAM.
While you don’t need to do so for this course, you can also easily install R and RStudio on your computer. For more information, see this page.
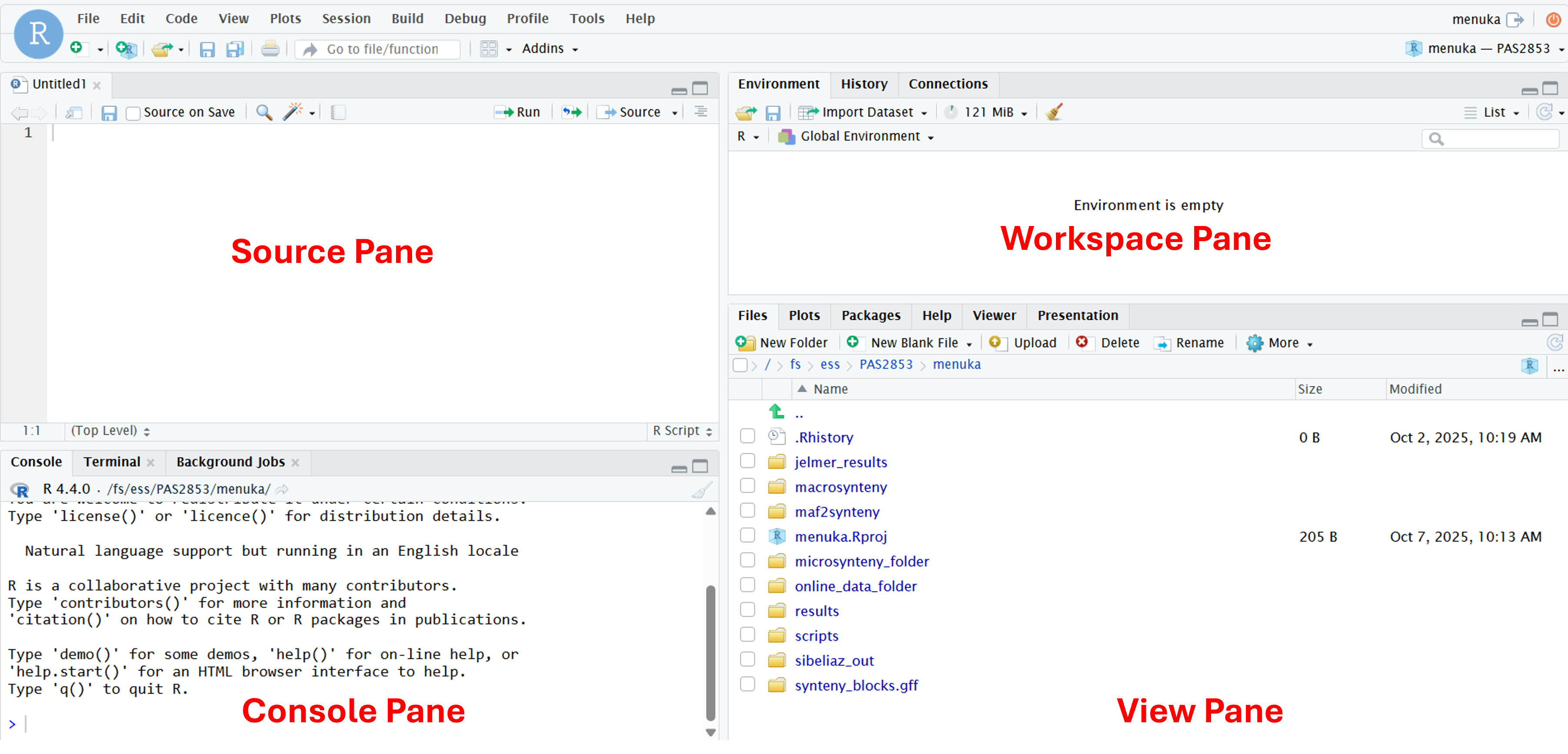
When you first open RStudio, you will be greeted by four panes, each of which have multiple tabs:
Top left: The editor/source pane, where you can open and edit documents like scripts
Bottom left: the default and most useful tab in this pane is your R “Console”, where you can type and execute R code
Top right: the default and most useful tab in this pane this is your “Environment”, which shows you all the objects that are active in your R environment
Bottom right: the default tab in this pane is “Files”, which is a file explorer starting in your initial working directory (more about that next). The additional tabs are also commonly used, such as those to show plots and help.
There are many things we can do in R: data manipulation, data visualization, statistical analysis, machine learning and many more. Here, to get some familiarity with working in R, we will start by simply using R as calculator.
To get used to typing and executing R code, we will simply use R as a calculator – type 5 + 5 in the console and press Enter:
R will print the result of the calculation, 10. The result is prefaced by [1], which is simply an index/count of output, which can be helpful when multiple numbers and elements are printed.
Some additional calculation examples:
Multiplication and division:
Exponents:
Parentheses can be used to change the default order of operations:
These codes will work with (like shown above) or without spaces between numbers and around operators like +. However, in general, it is good practice to leave space between the number to improve the readability of your code.
When using R as a calculator, the order of operations is the same as you would have learned back in school. From highest to lowest precedence:
(, )^ or ***/+-The > sign in your console is the R prompt, indicating that that R is ready for you to type something. In other words, this is equivalent to the $ Bash prompt that you are used to.
Also just like in Unix shell, when you are not seeing the > prompt, R is either busy (because you asked it to do a longer-running computation) or waiting for you to complete an incomplete command.
If you notice that your prompt turned into a +, you typed an incomplete command – for example:
+And once again like in the Unix shell, you can either try to finish the command or abort it by pressing Ctrl+C. In R, Esc also works to cancel – let’s practice the latter here.
In R, you can use comparison operators to compare numbers. When you do this, R will return a so-called logical (one of R’s “data types” – more about these later): either FALSE or TRUE. For example:
Greater than and less than:
Equal to or not equal to – don’t forget to use two equals signs == for the former:
Here are the most common comparison operators in R:
| Operator | Description | Example |
|---|---|---|
> |
Greater than | 5 > 6 returns FALSE |
< |
Less than | 5 < 6 returns TRUE |
== |
Equal to | 10 == 10 returns TRUE |
!= |
Not equal to | 10 != 10 returns FALSE |
>= |
Greater than or equal to | 5 >= 6 returns FALSE |
<= |
Less than or equal to | 6 <= 6 returns TRUE |
Find the natural log, log to the base 10, log to the base 2, square root and the natural antilog of 20.
Click to see a solution
To do almost anything in R, you will use “functions”, which are the equivalent of Unix commands. You can recognize function calls by the parentheses. For example:
The equivalent to the Shell’s pwd is getwd():
/fs/ess/PAS2880/usersInside those parentheses, you can provide arguments to a function. For example, using setwd(), the equivalent to the Shell’s cd:
we need to quote /fs/ess/PAS2880/users because anything that is not quoted in R has to be object
Note that unlike in the Shell, we need to quote the path! This is because in R, as you’ll see in more detail in a bit, anything that is not quoted is supposed to be an “object” like a variable.
One of the equivalents to the Shell’s echo is print():
The equivalent to the Shell’s mkdir is dir.create():
Now, we will look at the head() function, which shows the first six rows of a data frame or matrix. We will use it to visualize the data sets present in R.
The equivalent to the Shell’s head is head(), although it will by default show 6 lines/rows instead of 10:
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosairis come from?
R comes shipped with several built-in datasets, like iris. We didn’t need to (and shouldn’t) quote this, because it is a so-called object in R.
You can recognize function calls by parentheses. Inside those parentheses, you can provide arguments to a function. Arguments are the actual values you provided to run a function().
Arguments that are not object have to be in quotes in R
In R, you can choose to name the arguments in a function or just use their position, Named argument and positional argument. You can assign the values to argument, using the equal to sign =. When you assign values to the arguments, the order does not matter, R will know exactly which value goes where. For example, in head(n = 6, x = iris), the argument x is explicitly named so the function will run successfully. However, you can also write head(iris), and it gives the same output because R knows the first argument of head() is x. head(6, iris) will not produce any output because the arguments are called by position but are in the wrong order.
Consider the example below, where we pass the argument 6.53 to the function round():
In most functions, including round(), arguments have names that can be used. For example, the argument to round() representing the number to be rounded has the name x, so you can also use:
Naming arguments is helpful in particular to avoid confusion when you pass multiple arguments to a function. For example, round() accepts a second argument, which is the number of digits to round to:
You could choose not to name these arguments, but that is not as easy to understand:
Moreover, when you name arguments, you can give them in any order you want:
You can usually see which arguments a function takes by pausing after your type the function and its parenthesis, e.g. round():

Below, we’ll also see more extensive options to find out how a function works and what arguments it takes.
R has over 3,000 functions that serve many different purposes, and tens of thousands of additional functions are available in packages (add-ons). Below are a couple of examples continuing along the theme of using R as a calculator:
| Function | Description |
|---|---|
abs(x) |
Absolute value: e.g. abs(-5) returns 5 |
sqrt(x) |
Square root |
log(x) |
Natural logarithm |
log10(x) |
Common logarithm |
round(x, digits = n) |
Round: e.g round(3.475, digits = 2) returns 3.48 |
help() and ?The help() function and ? help operator in R offer access to documentation pages for R functions, data sets, and other objects. They provide access to both packages in the standard R distribution and contributed packages.
For example:
The output should appear in the Viewer tab of the bottom-left panel in RStudio and should look something like this:
You can assign one or more values to a so-called “object” with the assignment operator <-. A few examples:
[1] 250[1] 2.54In the shell, we’ve been calling these variables, and that terminology is also used for R objects like the above ones. However, R objects can take on many forms (we will talk about data structures in a bit), including more complicated things like tables. That’s why the general term object is used here.
To see the contents of an object, you can use the print() function as the equivalent of echo in the Unix shell:
However, in R you can also omit print() and simply type an object’s name:
Importantly, you can use objects as if you had typed their values directly:
Some pointers on object names:
Because R is case sensitive, length_inch is different from Length_Inch!
An object name cannot contain spaces — so for readability, you should separate words using:
length_inch (this is called “snake case”)wingspan.inchwingspanInch or WingspanInch (“camel case”)You will make things easier for yourself by naming objects in a consistent way, for instance, by always sticking to your favorite case style like “snake case.”
Object names can contain but cannot start with a number: x2 is valid but 2x is not.
Make object names descriptive yet not too long — this is not always easy!
Write the R code to assign the value 20 to the name num_1 and 15 to num_2.
What is sum of num_1 and num_2?
Assign the result of num_1 minus num_2 to a new object called difference. What is the value of difference?
Which of the following is a valid object name in R?
Click to see a solution
The valid object names are:
value_2
TotalValue
Object names cannot start with a number and cannot contain spaces (so a and c options are invalid).
R packages are basically add-ons or extensions to the functionality of the R language.
The function install.packages() installs an R package. This is a one-time operation, also at OSC (see below).
The function library() will load an R package in your current R session. Every time you want to use a package in R, you need to load it, just like everytime you want to use MS Word on your computer, you need to open it.
For example:
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ purrr 1.1.0
✔ forcats 1.0.0 ✔ readr 2.1.5
✔ ggplot2 3.5.2 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsIn RStudio server at OSC, packages will by default be installed into a standard location in your Home directory, so they can be accessed through time and regardless of what your working directory is, or what OSC project you are using1.
However, package installation is done separately for:
Different major versions of R (e.g. 4.1 versus 4.2, but not R 4.1.1 versus 4.1.5). Therefore, when you switch to a different R version, you will start over with package installation2.
Each OSC cluster, like Pitzer versus Cardinal.
As shown earlier, you can figure out what your working directory is with getwd(), and change it with setwd().
However, there is a better way of dealing with working directories, which is to avoid using setwd() altogether and to use RStudio Projects instead. This is similar to the Open Folder functionality of VS Code, but then a bit more formalized.
RSudio projects are useful for several reasons:
Automatically manage your working directory : RStudio treats project folder as a working directory, thus there is no need of manually setting working directory
Keep you work organized : Organize your project inside self-contained folders, which helps to manage, share and reproduce results
Ensure clean R sessions : Switching projects will restart R and remove all the objects and loaded packages
Easy to collaborate : All the paths in the RSudio projects will be in relative to the project folder. So, your code will work without modification.
To create a new RStudio Project inside your personal dir in /fs/ess/PAS2880:
File (top bar, below your browser’s address bar) > New ProjectExisting Directory.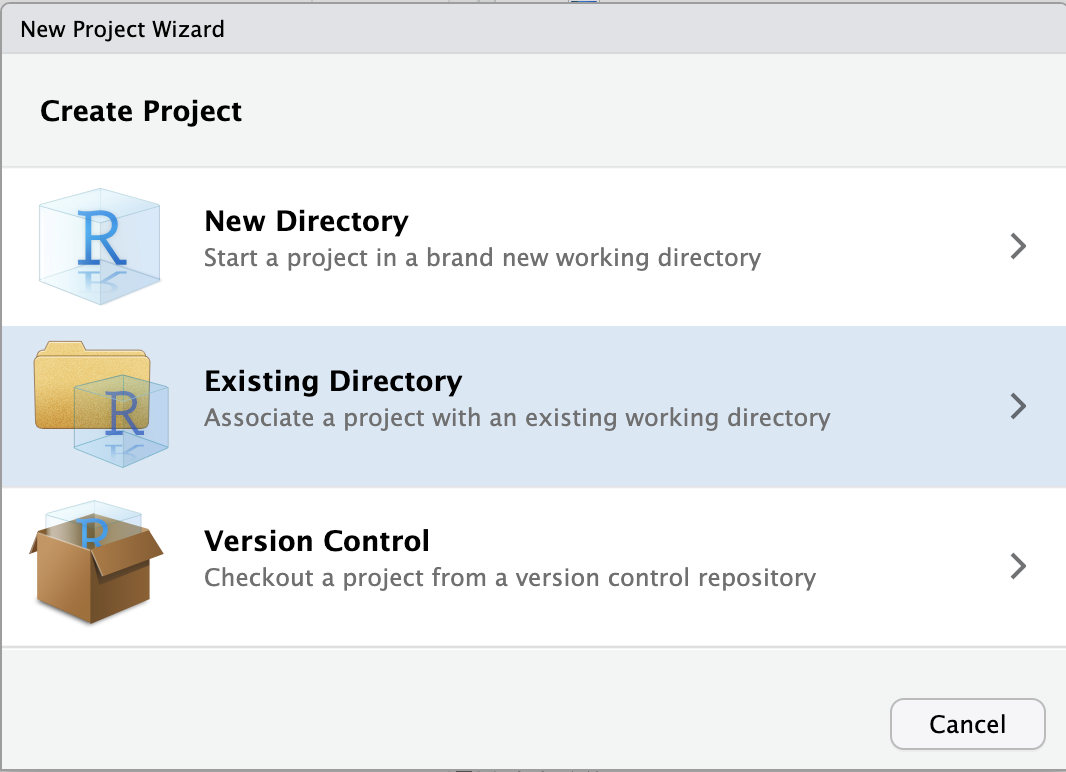
Browse... to select your personal dir.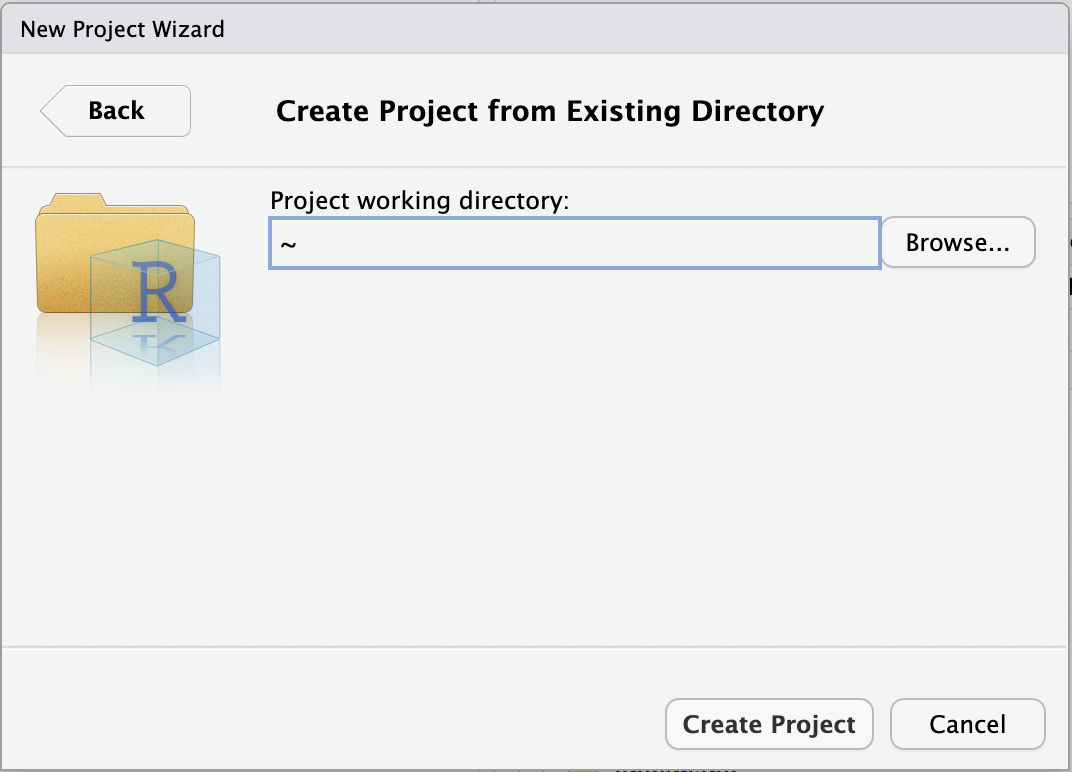
~), from which you can’t click your way to /fs/ess! Instead, you first have to click on the (very small!) ... highlighted in the screenshot below: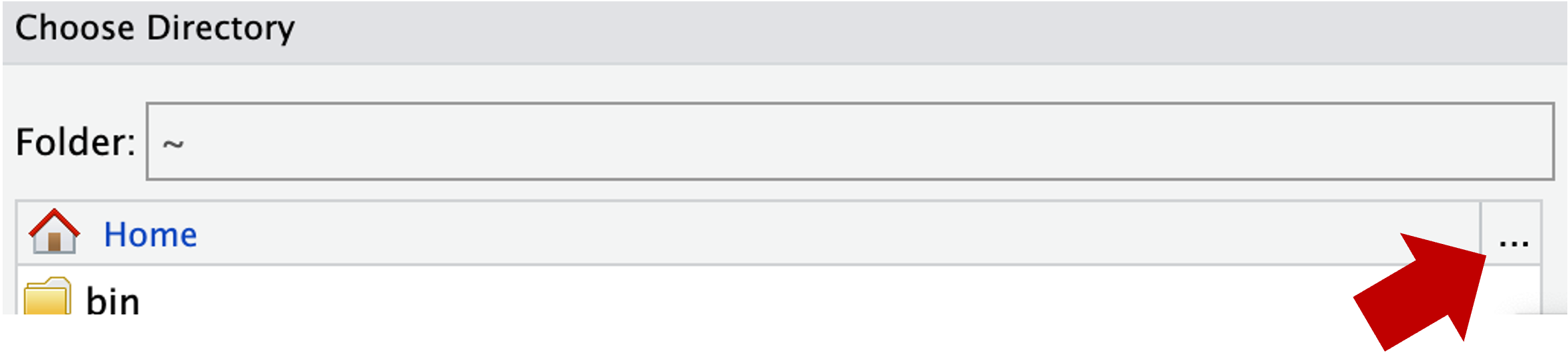
/fs/ess/PAS2880, e.g. as shown below, and click OK::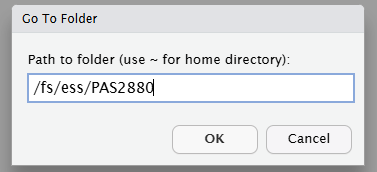
/fs/ess/PAS2880/users/$USER).Choose to pick your selected directory.Create Project.This will make R restart, which is by design. When R restarts, all objects are removed from the environment, packages are unloaded, and so on. This is a good idea whenever you switch to a different project, for example to avoid irreproducible carry-over effects.
Once you open a file, a new RStudio panel will appear in the top-left, where you can view and edit text files, most commonly R scripts (.R). An R script is a text file that contains R code.
Create and open a new R script by clicking File (top menu bar) > New File > R Script.
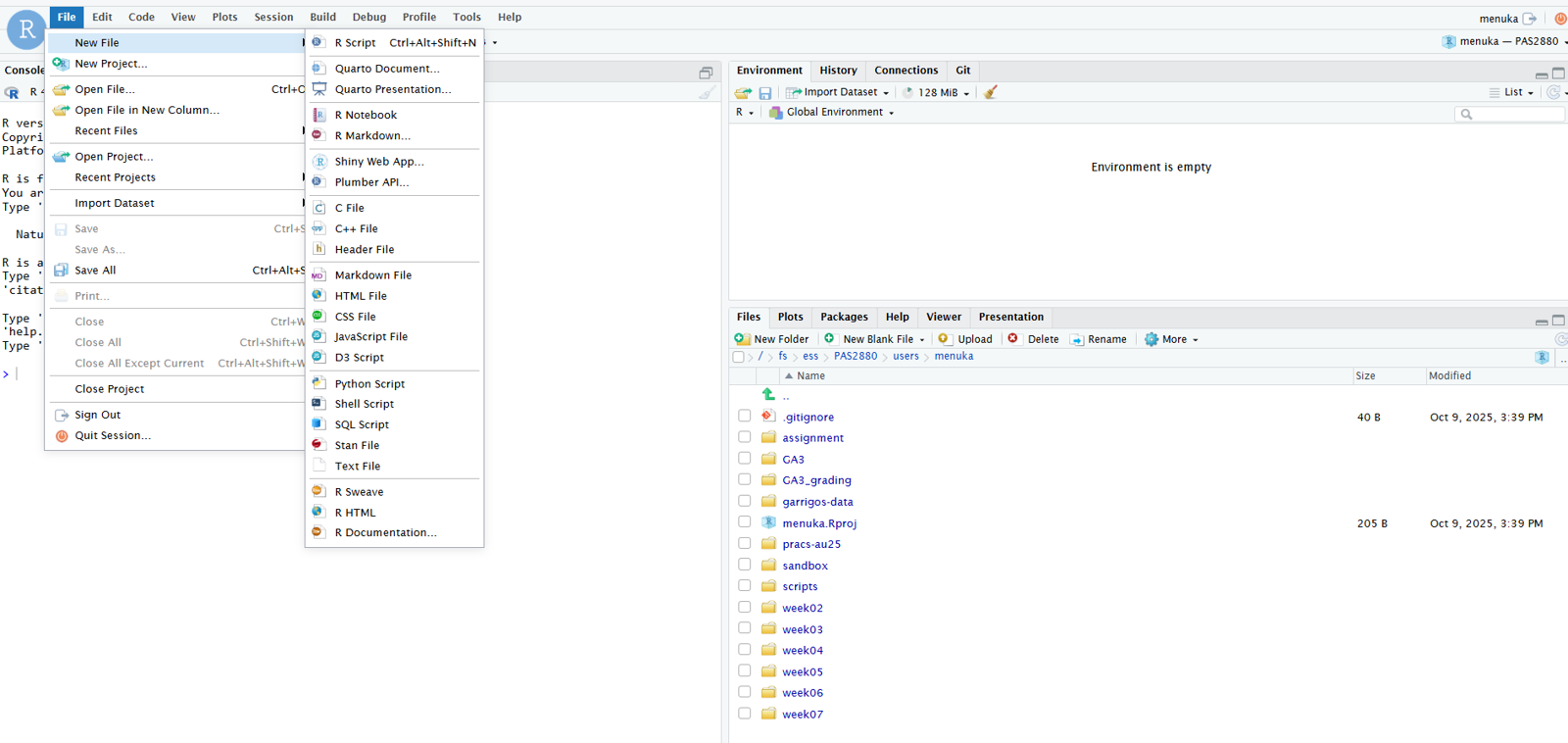
It’s a good idea to write and save most of your code in scripts, instead of directly in the R console. This helps to keep track of what you’ve been doing, especially in the longer run, and to re-run your code after modifying input data or one of the lines of code.
To send code from the editor to the console, where it will be executed by R, press Ctrl + Enter (or, on a Mac: Cmd + Enter) with the cursor anywhere in a line of code in the script.
Practice that after typing the following command in the script: